-
Quick start
-
API
-
-
-
-
-
-
- Dense
- PReLU 2D
- PReLU 3D
- PReLU 4D
- PReLU 5D
- AdditiveAttention
- Attention
- MutiHeadAttention
- Conv1D
- Conv2D
- Conv3D
- ConvLSTM1D
- ConvLSTM2D
- ConvLSTM3D
- Conv1DTranspose
- Conv2DTranspose
- Conv3DTranspose
- DepthwiseConv2D
- SeparableConv1D
- SeparableConv2D
- Embedding
- BatchNormalization
- LayerNormalization
- Bidirectional
- GRU
- LSTM
- SimpleRNN
- Show All Articles ( 12 ) Collapse Articles
-
- Dense
- PReLU 2D
- PReLU 3D
- PReLU 4D
- PReLU 5D
- AdditiveAttention
- Attention
- MultiHeadAttention
- Conv1D
- Conv2D
- Conv3D
- ConvLSTM1D
- ConvLSTM2D
- ConvLSTM3D
- Conv1DTranspose
- Conv2DTranspose
- Conv3DTranspose
- DepthwiseConv2D
- SeparableConv1D
- SeparableConv2D
- Embedding
- BatchNormalization
- LayerNormalization
- Bidirectional
- GRU
- LSTM
- SimpleRNN
- Show All Articles ( 12 ) Collapse Articles
-
-
- Dense
- AdditiveAttention
- Attention
- MultiHeadAttention
- BatchNormalization
- LayerNormalization
- Bidirectional
- GRU
- LSTM
- SimpleRNN
- Conv1D
- Conv2D
- Conv3D
- Conv1DTranspose
- Conv2DTranspose
- Conv3DTranspose
- ConvLSTM1D
- ConvLSTM2D
- ConvLSTM3D
- DepthwiseConv2D
- SeparableConv1D
- SeparableConv2D
- Embedding
- PReLU 2D
- PReLU 3D
- PReLU 4D
- PReLU 5D
- Show All Articles ( 12 ) Collapse Articles
-
-
- Dense
- Embedding
- AdditiveAttention
- Attention
- MultiHeadAttention
- Conv1D
- Conv2D
- Conv3D
- ConvLSTM1D
- ConvLSTM2D
- ConvLSTM3D
- Conv1DTranspose
- Conv2DTranspose
- Conv3DTranspose
- DepthwiseConv2D
- SeparableConv1D
- SeparableConv2D
- BatchNormalization
- LayerNormalization
- PReLU 2D
- PReLU 3D
- PReLU 4D
- PReLU 5D
- Bidirectional
- GRU
- LSTM
- RNN (GRU)
- RNN (LSTM)
- RNN (SimpleRNN)
- SimpleRNN
- Show All Articles ( 15 ) Collapse Articles
-
- Dense
- Embedding
- AdditiveAttention
- Attention
- MultiHeadAttention
- Conv1D
- Conv2D
- Conv3D
- ConvLSTM1D
- ConvLSTM2D
- ConvLSTM3D
- Conv1DTranspose
- Conv2DTranspose
- Conv3DTranspose
- DepthwiseConv2D
- SeparableConv1D
- SeparableConv2D
- BatchNormalization
- LayerNormalization
- PReLU 2D
- PReLU 3D
- PReLU 4D
- PReLU 5D
- Bidirectional
- GRU
- LSTM
- RNN (GRU)
- RNN (LSTM)
- RNN (SimpleRNN)
- SimpleRNN
- Show All Articles ( 15 ) Collapse Articles
-
-
-
- Dense
- Embedding
- AdditiveAttention
- Attention
- MultiHeadAttention
- Conv1D
- Conv2D
- Conv3D
- ConvLSTM1D
- ConvLSTM2D
- ConvLSTM3D
- Conv1DTranspose
- Conv2DTranspose
- Conv3DTranspose
- DepthwiseConv2D
- SeparableConv1D
- SeparableConv2D
- BatchNormalization
- LayerNormalization
- PReLU 2D
- PReLU 3D
- PReLU 4D
- PReLU 5D
- Bidirectional
- GRU
- LSTM
- RNN (GRU)
- RNN (LSTM)
- RNN (SimpleRNN)
- SimpleRNN
- Show All Articles ( 15 ) Collapse Articles
-
- Dense
- Embedding
- AdditiveAttention
- Attention
- MultiHeadAttention
- Conv1D
- Conv2D
- Conv3D
- ConvLSTM1D
- ConvLSTM2D
- ConvLSTM3D
- Conv1DTranspose
- Conv2DTranspose
- Conv3DTranspose
- DepthwiseConv2D
- SeparableConv1D
- SeparableConv2D
- BatchNormalization
- LayerNormalization
- PReLU 2D
- PReLU 3D
- PReLU 4D
- PReLU 5D
- Bidirectional
- GRU
- LSTM
- RNN (GRU)
- RNN (LSTM)
- RNN (SimpleRNN)
- SimpleRNN
- Show All Articles ( 15 ) Collapse Articles
-
-
-
-
- Add
- AdditiveAttention
- AlphaDropout
- Attention
- Average
- AvgPool1D
- AvgPool2D
- AvgPool3D
- BatchNormalization
- Bidirectional
- Concatenate
- Conv1D
- Conv1DTranspose
- Conv2D
- Conv2DTranspose
- Conv3D
- Conv3DTranspose
- ConvLSTM1D
- ConvLSTM2D
- ConvLSTM3D
- Cropping1D
- Cropping2D
- Cropping3D
- Dense
- DepthwiseConv2D
- Dropout
- Embedding
- Flatten
- GaussianDropout
- GaussianNoise
- GlobalAvgPool1D
- GlobalAvgPool2D
- GlobalAvgPool3D
- GlobalMaxPool1D
- GlobalMaxPool2D
- GlobalMaxPool3D
- GRU
- Input
- LayerNormalization
- LSTM
- MaxPool1D
- MaxPool2D
- MaxPool3D
- MultiHeadAttention
- Multiply
- Permute3D
- Reshape
- RNN
- SeparableConv1D
- SeparableConv2D
- SimpleRNN
- SpatialDropout
- Substract
- TimeDistributed
- UpSampling1D
- UpSampling2D
- UpSampling3D
- ZeroPadding1D
- ZeroPadding2D
- ZeroPadding3D
- Show All Articles ( 45 ) Collapse Articles
-
- AlphaDropout
- AvgPool1D
- AvgPool2D
- AvgPool3D
- BatchNormalization
- Bidirectional
- Conv1D
- Conv1DTranspose
- Conv2D
- Conv2DTranspose
- Conv3D
- Conv3DTranspose
- Cropping1D
- Cropping2D
- Cropping3D
- Dense
- DepthwiseConv2D
- Dropout
- Embedding
- Flatten
- GaussianDropout
- GaussianNoise
- GlobalAvgPool1D
- GlobalAvgPool2D
- GlobalAvgPool3D
- GlobalMaxPool1D
- GlobalMaxPool2D
- GlobalMaxPool3D
- GRU
- LayerNormalization
- LSTM
- MaxPool1D
- MaxPool2D
- MaxPool3D
- Permute3D
- Reshape
- RNN
- SeparableConv1D
- SeparableConv2D
- SimpleRNN
- SpatialDropout
- UpSampling1D
- UpSampling2D
- UpSampling3D
- ZeroPadding1D
- ZeroPadding2D
- ZeroPadding3D
- Show All Articles ( 32 ) Collapse Articles
-
-
-
- Resume
- Accuracy
- BinaryAccuracy
- BinaryCrossentropy
- BinaryIoU
- CategoricalAccuracy
- CategoricalCrossentropy
- CategoricalHinge
- CosineSimilarity
- FalseNegatives
- FalsePositives
- Hinge
- Huber
- IoU
- KLDivergence
- LogCoshError
- Mean
- MeanAbsoluteError
- MeanAbsolutePercentageError
- MeanIoU
- MeanRelativeError
- MeanSquaredError
- MeanSquaredLogarithmicError
- MeanTensor
- OneHotIoU
- OneHotMeanIoU
- Poisson
- Precision
- PrecisionAtRecall
- Recall
- RecallAtPrecision
- RootMeanSquaredError
- SensitivityAtSpecificity
- SparseCategoricalAccuracy
- SparseCategoricalCrossentropy
- SparseTopKCategoricalAccuracy
- Specificity
- SpecificityAtSensitivity
- SquaredHinge
- Sum
- TopKCategoricalAccuracy
- TrueNegatives
- TruePositives
- Show All Articles ( 28 ) Collapse Articles
-
- Resume
- Constant
- GlorotNormal
- GlorotUniform
- HeNormal
- HeUniform
- Identity
- LecunNormal
- LecunUniform
- Ones
- Orthogonal
- RandomNormal
- RandomUnifom
- TruncatedNormal
- VarianceScaling
- Zeros
- Show All Articles ( 1 ) Collapse Articles
-
ConvLSTM2DCell
Description
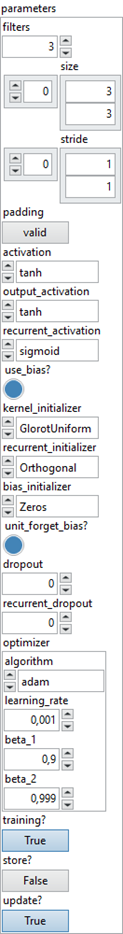
Define the cell convolution lstm 2D layer according to its parameters. To be used for the ConvLSTM2D layer. Type : polymorphic.
![]()
Input parameters
![]() parameters : layer parameters.
parameters : layer parameters.
![]() filters : integer, the dimensionality of the output space.
filters : integer, the dimensionality of the output space.
Default value “3”.![]() size : array integer, specify the dimensions of the convolution window.
size : array integer, specify the dimensions of the convolution window.
Default value “[3,3]”.![]() stride : array integer, specify the strides of the convolution.
stride : array integer, specify the strides of the convolution.
Default value “[1,1]”.![]() padding : boolean, False = “valid” means no padding. True = “same” results in padding with zeros evenly to the left/right or up/down of the input such that output has the same height/width dimension as the input.
padding : boolean, False = “valid” means no padding. True = “same” results in padding with zeros evenly to the left/right or up/down of the input such that output has the same height/width dimension as the input.
Default value “False”.![]() activation : enum, activation function to use.
activation : enum, activation function to use.
Default value “tanh”.![]() output_activation : enum, activation function to use.
output_activation : enum, activation function to use.
Default value “tanh”.![]() recurrent_activation : enum, activation function to use for the recurrent step.
recurrent_activation : enum, activation function to use for the recurrent step.
Default value “sigmoid”.![]() use_bias? : boolean, whether the layer uses a bias vector.
use_bias? : boolean, whether the layer uses a bias vector.
Default value “True”.![]() kernel_initializer : enum, initializer for the kernel weights matrix, used for the linear transformation of the inputs.
kernel_initializer : enum, initializer for the kernel weights matrix, used for the linear transformation of the inputs.
Default value “glorot_uniform”.![]() recurrent_initializer : enum, initializer for the recurrent_kernel weights matrix, used for the linear transformation of the recurrent state.
recurrent_initializer : enum, initializer for the recurrent_kernel weights matrix, used for the linear transformation of the recurrent state.
Default value “orthogonal”.![]() bias_initializer : enum, initializer for the bias vector.
bias_initializer : enum, initializer for the bias vector.
Default value “zero”.![]() unit_forget_bias? : boolean, if True, add 1 to the bias of the forget gate at initialization.
unit_forget_bias? : boolean, if True, add 1 to the bias of the forget gate at initialization.
Default value “True”.![]() dropout : float, fraction of the units to drop for the linear transformation of the inputs.
dropout : float, fraction of the units to drop for the linear transformation of the inputs.
Default value “0.0”.![]() recurrent_dropout : float, fraction of the units to drop for the linear transformation of the recurrent state.
recurrent_dropout : float, fraction of the units to drop for the linear transformation of the recurrent state.
Default value “0.0”.![]() optimizer :
optimizer :
![]() algorithm : enum, (name of optimizer) for optimizer instance.
algorithm : enum, (name of optimizer) for optimizer instance.
Default value “adam”.![]() learning_rate : float, define the learning rate to use.
learning_rate : float, define the learning rate to use.
Default value “0.001”.![]() beta_1 : float, define the exponential decay rate for the 1st moment estimates.
beta_1 : float, define the exponential decay rate for the 1st moment estimates.
Default value “0.9”.![]() beta_2 : float, define the exponential decay rate for the 2nd moment estimates.
beta_2 : float, define the exponential decay rate for the 2nd moment estimates.
Default value “0.999”.
![]() training? : boolean, whether the layer is in training mode (can store data for backward).
training? : boolean, whether the layer is in training mode (can store data for backward).
Default value “True”.![]() store? : boolean, whether the layer stores the last iteration gradient (accessible via the “get_gradients” function).
store? : boolean, whether the layer stores the last iteration gradient (accessible via the “get_gradients” function).
Default value “False”.![]() update? : boolean, whether the layer’s variables should be updated during backward. Equivalent to freeze the layer.
update? : boolean, whether the layer’s variables should be updated during backward. Equivalent to freeze the layer.
Default value “True”.
Output parameters
![]() ConvLSTM2DCell out : layer cell convolution lstm 2d architecture
ConvLSTM2DCell out : layer cell convolution lstm 2d architecture
Example
All these exemples are snippets PNG, you can drop these Snippet onto the block diagram and get the depicted code added to your VI (Do not forget to install HAIBAL library to run it).
ConvLSTM2D cell inside ConvLSTM2D layer
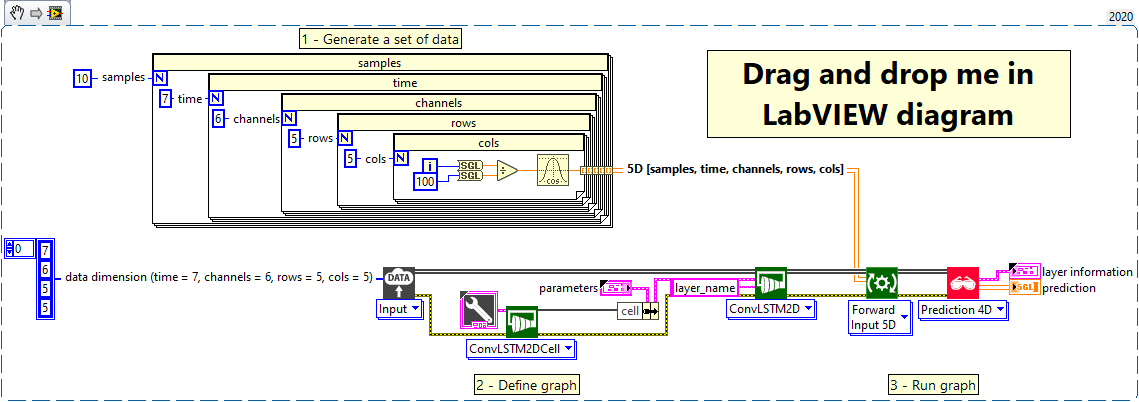
1 – Generate a set of data
We generate an array of data of type single and shape [samples = 10, time = 7, channels = 6, rows = 5, cols = 5]
2 – Define graph
First, we define the first layer of the graph which is an Input layer (explicit input layer method). This layer is setup as an input array shaped [time = 7, channels = 6, rows = 5, cols = 5].
Next, we configure the “cell” parameter of the ConvLSTM2D layer with the ConvLSTM2D cell.
Finally, we add to the graph the ConvLSTM2D layer.
3 – Run graph
We call the forward method and retrieve the result with the “Prediction 4D” method.
This method returns two variables, the first one is the layer information (cluster composed of the layer name, the graph index and the shape of the output layer) and the second one is the prediction with a shape of [samples, filters, new_rows, new_cols].
The output dimension depends on the parameters “return-sequences” refer to the chapter “Dimension” of ConvLSTM2D add to graph documentation.